Deep learning has transformed medical image analysis by delivering clinically meaningful results on challenging problems like prostate cancer detection and lung screening. In pathology, industry is making significant investments to develop deep learning tools for diagnostic use in clinical labs. FDA approval of whole-slide digital pathology images (WSIs) for use in primary diagnosis is further increasing interest, adoption, and investment in this technology. Judgments made by pathologists are the basis for the treatment of many diseases, yet interobserver variability among pathologists is significant, and errors can lead to overtreatment or even treatment of healthy patients. Pathology is also facing workforce issues as demand for pathologist services is outpacing growth of trained pathologists. Computational pathology tools based on deep learning can help address these problems by providing reproducible diagnoses, performing ”second reads” for human pathologists, automating tasks to improve pathologist efficiency, and helping general pathologists evaluate challenging cases. GPU accel-erators have played a significant role in advancing deep learning methods to build computational pathology tools, with machine learning frameworks (MLFs) like Pytorch and Tensorflow providing researchers with abstractions to quickly develop models that utilize GPUs. Evolution of GPUs and MLFs has been driven by analysis of small images, and so these tools cannot be easily applied directly WSIs or other large medical images like three dimensional MRI or CT. Adapting medical imaging problems to small image paradigms supported by GPUs and MLFs leads to suboptimal performance and increased implementation effort and complexity. More recent approaches that use streaming or ”unified memory” allow direct analysis of entire WSIs and have demonstrated performance advantages. These approaches can be slow, complex to implement, and are highly specific to a choice of network architecture which limits exploration and development of new architectures. More general-purpose, efficient, and user-friendly frameworks are required to allow the development of WSI scale deep learning.
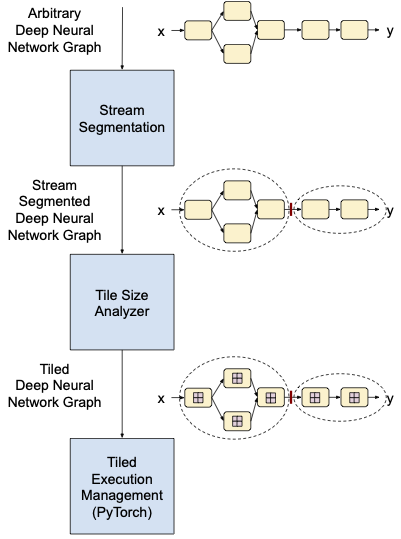
This tools will develop techniques to automatically map deep learning networks implemented in common MLF architectures to one or more GPUs for arbitrarily large input images and activation layers. The proposed software will include a performance modeler to estimate the runtime of a given network on available GPU accelerators. These strategies will enable a new paradigm in deep learning for medical images, allowing the developmentof novel networks that are purpose-built for medical applications. Developers will be able to rapidly create andevaluate these networks using familiar MLF packages. This project will provide approaches to overcome GPU memory bottlenecks, a scheduler to map the network to available GPUs, integration with common MLFs, and demonstration using computational pathology use cases.